Building an AI Knowledge Assistant
After completing the 5-Day Gen AI Intensive Course and diving into Introduction to LangGraph, I embarked on building a personal AI Knowledge Assistant. This post documents my exploration of multi-agent processing using LangGraph.
Unlike my earlier posts on Coding with CrewAI and Multi-agent Conservation with Autogen, this effort focuses on creating a sophisticated multi-agent Retrieval-Augmented Generation (RAG) system using LangGraph.
Getting Started
The Windsurf Editor
For this project, I utilized the innovative Windsurf Editor, an agent-first IDE. Using the initial prompt:
I want to build a multi-agent RAG with LangGraph using local Llama3.
I began crafting my AI Knowledge Assistant. Below is a screenshot of the initial code:
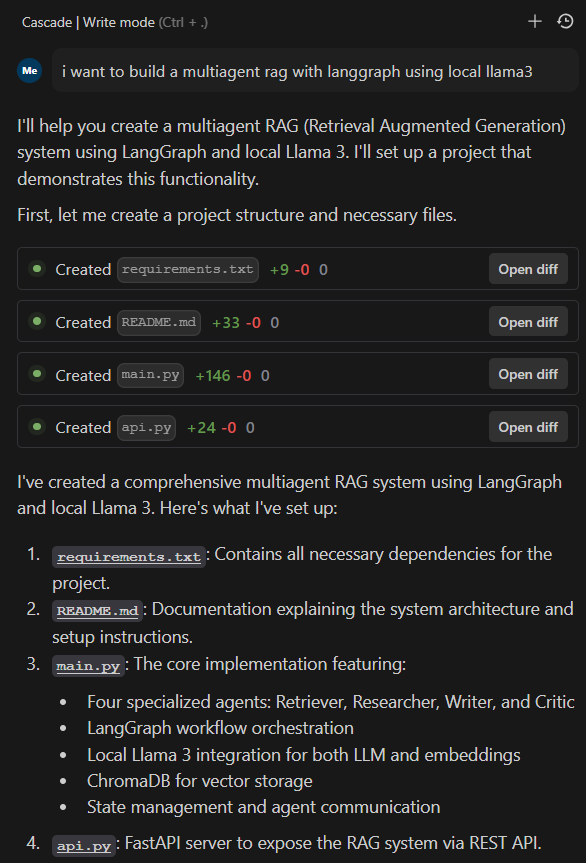
Setting Up the Environment
Running Ollama in Docker
To power this system, I deployed the Llama3 model using Ollama in a Docker container:
docker run -d --gpus=all -v $PWD/ollama:/root/.ollama -p 11434:11434 --name ollama ollama/ollama
Then, I started the Llama3 model:
docker exec -it ollama ollama run llama3
Alternatively, Ollama can be installed natively on Windows, and the model can be run directly:
ollama run llama3
Building the AI Knowledge Assistant
Initial Code Structure
Below is the initial code for main.py. It leverages LangGraph to coordinate multiple agents for query processing.
from typing import Dict, TypedDict, Sequence
from langchain_core.messages import BaseMessage, HumanMessage, AIMessage
from langgraph.graph import StateGraph
from langchain_core.prompts import ChatPromptTemplate
from langchain_core.output_parsers import StrOutputParser
from langchain_ollama import OllamaLLM, OllamaEmbeddings
from langchain_chroma import Chroma
import os
from dotenv import load_dotenv
from IPython.display import Image, display
load_dotenv()
# Initialize LLM with Ollama
OLLAMA_BASE_URL = os.getenv("OLLAMA_BASE_URL", "http://host.docker.internal:11434")
MODEL_NAME = os.getenv("OLLAMA_MODEL", "llama3")
llm = OllamaLLM(
base_url=OLLAMA_BASE_URL,
model=MODEL_NAME,
temperature=0.75,
)
# Initialize embeddings with Ollama
embeddings = OllamaEmbeddings(
base_url=OLLAMA_BASE_URL,
model=MODEL_NAME,
)
# Initialize vector store
vectorstore = Chroma(
persist_directory="./chroma_db",
embedding_function=embeddings,
collection_name="rag_collection"
)
class AgentState(TypedDict):
messages: Sequence[BaseMessage]
current_step: str
context: str
research_summary: str
final_answer: str
def retriever_agent(state: AgentState) -> Dict:
"""Agent responsible for retrieving relevant documents."""
query = state["messages"][-1].content
# Search vector store
docs = vectorstore.similarity_search(query, k=3)
context = "\n".join([doc.page_content for doc in docs])
return {
**state,
"context": context,
"current_step": "researcher"
}
def researcher_agent(state: AgentState) -> Dict:
"""Agent responsible for analyzing retrieved documents."""
researcher_prompt = ChatPromptTemplate.from_template(
"""Based on the following context and question, provide a detailed analysis:
Context: {context}
Question: {question}
Analysis:"""
)
chain = researcher_prompt | llm | StrOutputParser()
research_summary = chain.invoke({
"context": state["context"],
"question": state["messages"][-1].content
})
return {
**state,
"research_summary": research_summary,
"current_step": "writer"
}
def writer_agent(state: AgentState) -> Dict:
"""Agent responsible for composing the final response."""
writer_prompt = ChatPromptTemplate.from_template(
"""Based on the research summary, compose a clear and concise response:
Research Summary: {research_summary}
Original Question: {question}
Response:"""
)
chain = writer_prompt | llm | StrOutputParser()
final_answer = chain.invoke({
"research_summary": state["research_summary"],
"question": state["messages"][-1].content
})
return {
**state,
"final_answer": final_answer,
"current_step": "critic"
}
def critic_agent(state: AgentState) -> Dict:
"""Agent responsible for reviewing and refining the response."""
critic_prompt = ChatPromptTemplate.from_template(
"""Review and improve the following response if necessary:
Context: {context}
Original Question: {question}
Current Response: {response}
Improved Response:"""
)
chain = critic_prompt | llm | StrOutputParser()
improved_answer = chain.invoke({
"context": state["context"],
"question": state["messages"][-1].content,
"response": state["final_answer"]
})
new_messages = list(state["messages"])
new_messages.append(AIMessage(content=improved_answer))
return {
**state,
"messages": new_messages
}
# Create the graph
workflow = StateGraph(AgentState)
# Add nodes
workflow.add_node("retriever", retriever_agent)
workflow.add_node("researcher", researcher_agent)
workflow.add_node("writer", writer_agent)
workflow.add_node("critic", critic_agent)
# Set the entrypoint
workflow.set_entry_point("retriever")
# Add edges
workflow.add_edge("retriever", "researcher")
workflow.add_edge("researcher", "writer")
workflow.add_edge("writer", "critic")
workflow.add_edge("critic", END)
# Compile the graph
chain = workflow.compile()
# Show
display(Image(chain.get_graph(xray=True).draw_mermaid_png()))
def process_query(query: str) -> str:
"""Process a query through the multi-agent system."""
result = chain.invoke({
"messages": [HumanMessage(content=query)],
"current_step": "retriever",
"context": "",
"research_summary": "",
"final_answer": ""
})
return result["messages"][-1].content
if __name__ == "__main__":
# Example usage
query = "What are the benefits of using a Retrival Augmented Generation system?"
response = process_query(query)
print(f"Query: {query}")
print(f"Response: {response}")
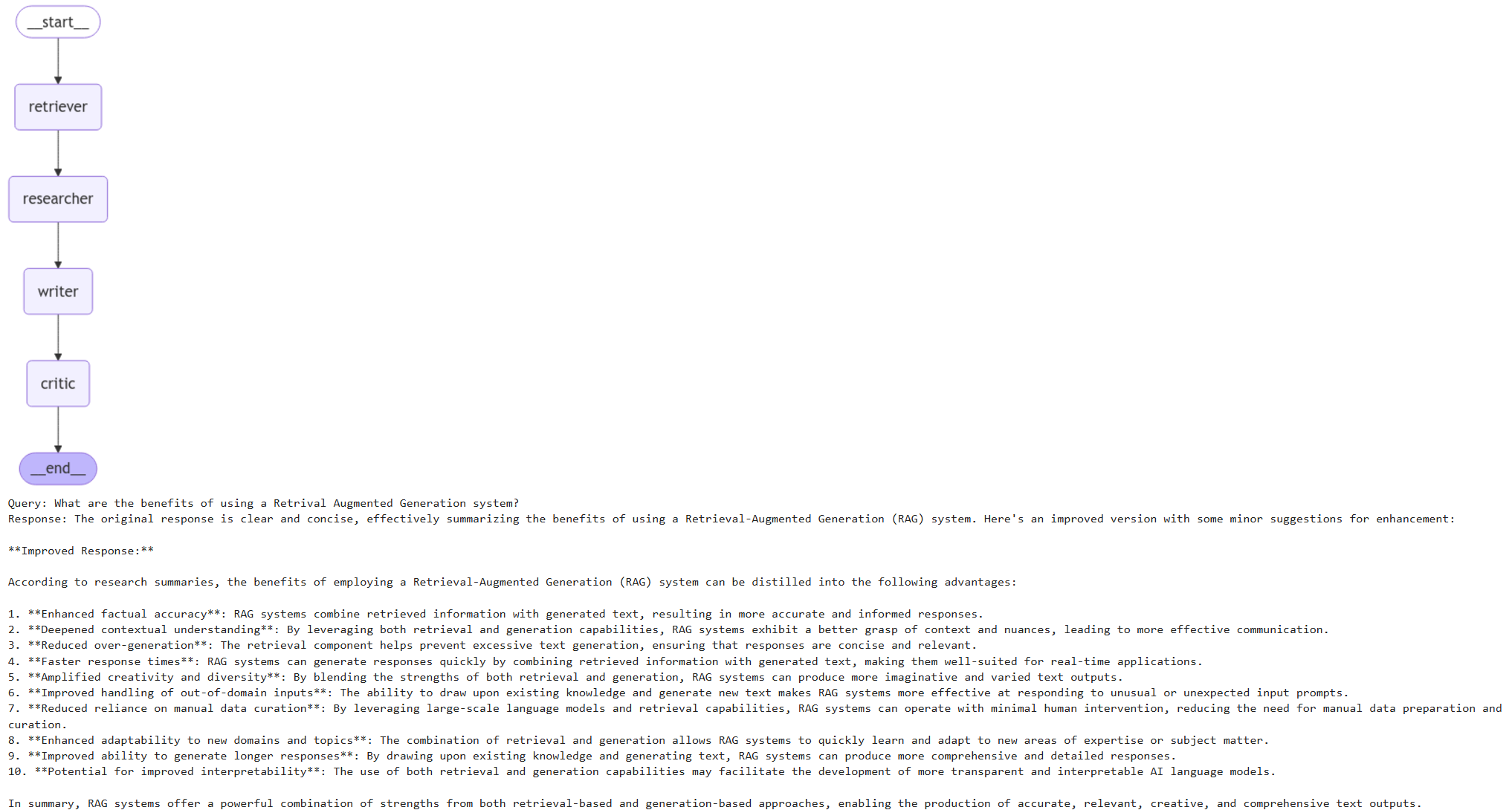
This framework coordinates multiple agents—Retriever, Researcher, Writer, and Critic—to collaboratively process and answer queries.
Sample Workflow
The workflow begins with the Retriever Agent fetching relevant documents using a vector store and proceeds through subsequent agents to analyze, compose, and refine responses. Below is an example snippet for the Retriever Agent:
def retriever_agent(state: AgentState) -> AgentState:
"""Retriever agent that finds relevant documents."""
print("\n=== Retriever Agent ===")
print(f"Input state: {state}")
messages = state["messages"]
query = messages[-1].content
print(f"Processing query: {query}")
# Get relevant documents
docs = vectorstore.similarity_search(query, k=3)
context = "\n\n".join([doc.page_content for doc in docs])
print(f"Found {len(docs)} documents")
# Initialize agent outputs if not present
if "agent_outputs" not in state:
state["agent_outputs"] = {}
# Format markdown response
response = {
"retriever": f"""<details open>
<summary>### 📚 Retrieved Documents</summary>
Found {len(docs)} relevant documents.
<details>
<summary>Document Content</summary>
\```
{context}
\```
</details>
</details>
"""
}
# Update state while preserving existing fields
state["current_step"] = "researcher"
state["context"] = context
state["query"] = query
state["agent_outputs"]["retriever"] = response["retriever"]
print(f"Output state: {state}")
return state
Data Ingestion
Document Processor
To make the system versatile, I developed processor.py for ingesting documents into the vector store. It supports various file formats like PDFs, text, and markdown:
from typing import Dict, List, Optional
import os
from pathlib import Path
from langchain_community.document_loaders import (
PyPDFLoader,
TextLoader,
UnstructuredMarkdownLoader,
DirectoryLoader
)
from langchain.text_splitter import RecursiveCharacterTextSplitter
from datetime import datetime
from main import vectorstore
class Processor:
"""A processor that ingests various types of documents into a vector store for later retrieval."""
SUPPORTED_EXTENSIONS = {
'.pdf': PyPDFLoader,
'.txt': TextLoader,
'.md': UnstructuredMarkdownLoader
}
def __init__(self):
self.text_splitter = RecursiveCharacterTextSplitter(
chunk_size=1000,
chunk_overlap=200
)
def process_document(self, file_path: str, document_type: Optional[str] = None) -> bool:
"""Process a single document file into the vector store."""
try:
path = Path(file_path)
if not path.exists():
raise FileNotFoundError(f"File not found: {file_path}")
# Get file extension and appropriate loader
ext = path.suffix.lower()
if ext not in self.SUPPORTED_EXTENSIONS:
raise ValueError(f"Unsupported file type: {ext}. Supported types: {list(self.SUPPORTED_EXTENSIONS.keys())}")
print(f"Processing {path.name}...")
loader_class = self.SUPPORTED_EXTENSIONS[ext]
loader = loader_class(str(path))
# Load and process document
documents = loader.load()
# Add metadata
for doc in documents:
doc.metadata.update({
'source': str(path),
'filename': path.name,
'type': document_type or 'general',
'date_processed': datetime.now().isoformat(),
})
# Split documents into chunks
splits = self.text_splitter.split_documents(documents)
# Add to vector store
vectorstore.add_documents(splits)
print(f"Successfully processed {path.name} ({len(splits)} chunks created)")
return True
except Exception as e:
print(f"Error processing document {file_path}: {str(e)}")
return False
def process_directory(self, directory_path: str, document_type: Optional[str] = None) -> Dict[str, int]:
"""Process all supported documents in a directory."""
stats = {
'total': 0,
'successful': 0,
'failed': 0
}
try:
path = Path(directory_path)
if not path.exists() or not path.is_dir():
raise NotADirectoryError(f"Directory not found: {directory_path}")
# Process each supported file in the directory
for ext, _ in self.SUPPORTED_EXTENSIONS.items():
files = list(path.glob(f"**/*{ext}"))
stats['total'] += len(files)
for file_path in files:
if self.process_document(str(file_path), document_type):
stats['successful'] += 1
else:
stats['failed'] += 1
except Exception as e:
print(f"Error processing directory {directory_path}: {str(e)}")
return stats
def main():
processor = Processor()
while True:
print("\n=== Knowledge Base ===")
print("1. Process single document")
print("2. Process directory")
print("3. Exit")
choice = input("\nEnter your choice (1-3): ")
if choice == "1":
file_path = input("Enter the path to your document: ")
doc_type = input("Enter document type (press Enter for 'general'): ")
if processor.process_document(file_path, doc_type or None):
print("Document processed successfully!")
elif choice == "2":
dir_path = input("Enter the directory path: ")
doc_type = input("Enter document type (press Enter for 'general'): ")
stats = processor.process_directory(dir_path, doc_type or None)
print(f"\nProcessing complete:")
print(f"Total files: {stats['total']}")
print(f"Successfully processed: {stats['successful']}")
print(f"Failed: {stats['failed']}")
elif choice == "3":
break
else:
print("Invalid choice. Please try again.")
if __name__ == "__main__":
main()
This modular ingestion pipeline ensures that the AI Knowledge Assistant has access to relevant and up-to-date knowledge.
Ingesting Data
To ingest data for the AI Knowledge Assistant, let’s start with a sample knowledge base, such as the Microservices Design Patterns. This step involves running the data ingestion script:
python3 processor.py
Once executed, the script processes the document and stores the extracted knowledge in a format optimized for querying.

Running AI Knowledge Assistant
With the data ingested, we can now start the AI Knowledge Assistant by running the following command:
python3 api.py
# Sample result
# INFO: Started server process [729]
# INFO: Waiting for application startup.
# INFO: Application startup complete.
# INFO: Uvicorn running on http://0.0.0.0:8000 (Press CTRL+C to quit)
First question
Let’s test the assistant by asking a question:
How does the CQRS pattern improve scalability and performance in application design?
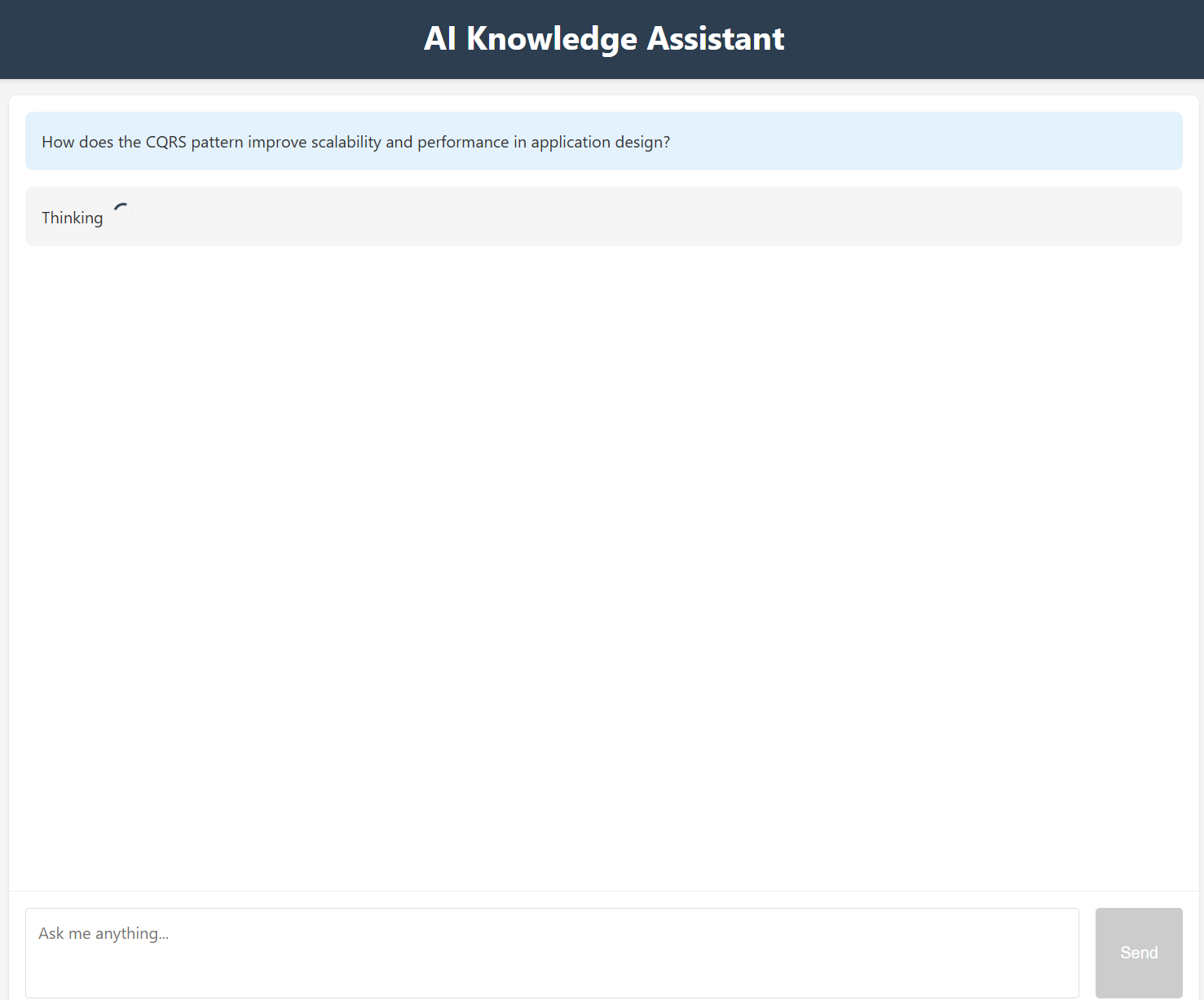
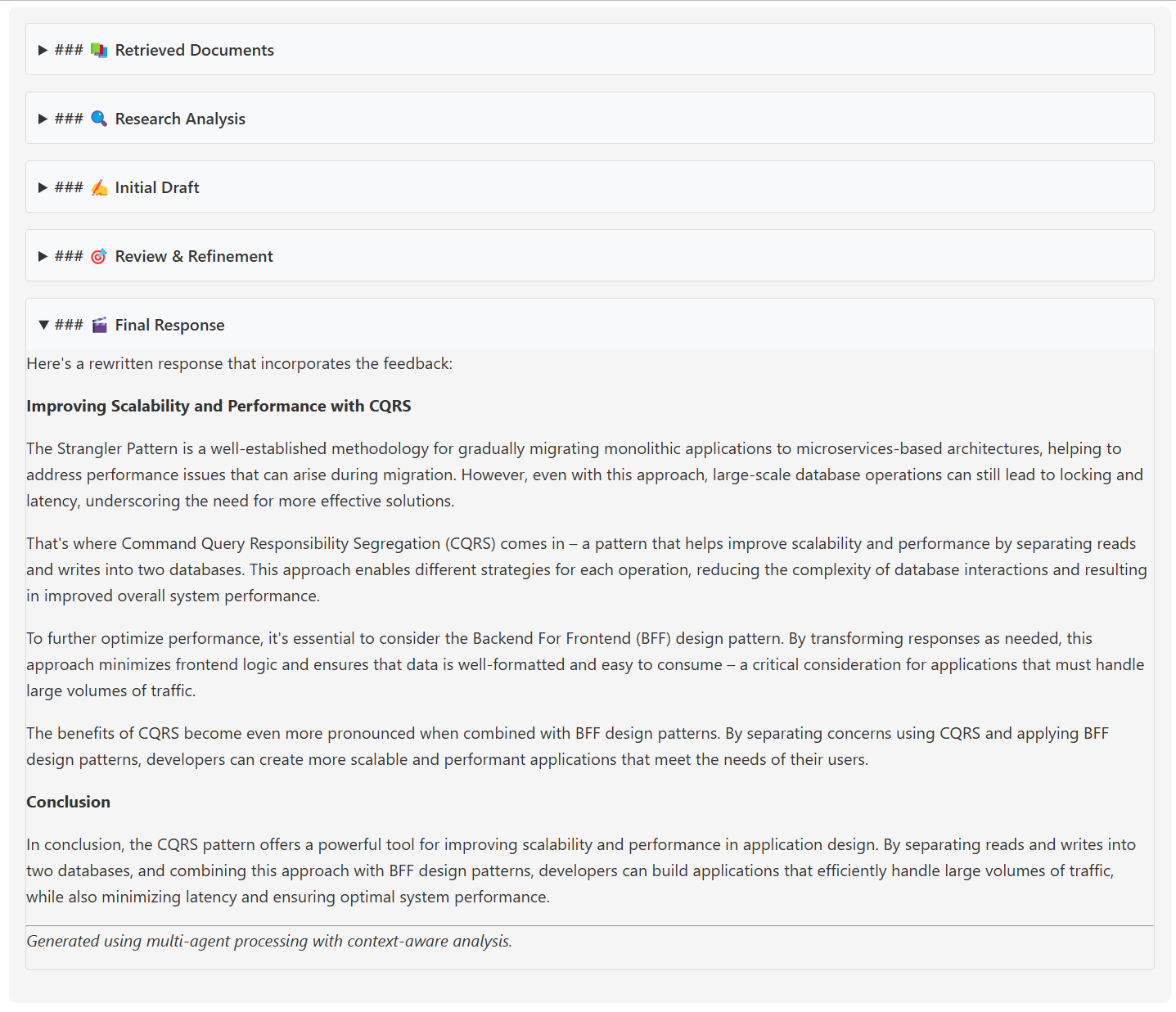
Here’s an example of the response from the retriever-agent as it fetches relevant knowledge:
The final synthesized response provided by the AI Knowledge Assistant looks like this:
Code Repository
The full implementation of this AI Knowledge Assistant is available on GitHub. Explore the code, and try it out yourself at https://github.com/seehiong/ai-knowledge-assistant.